Accurate Molecular Detection of S. aureus
Diagnostic Accuracy of Presurgical Staphylococcus aureus PCR Assay Compared with Culture and Post-PCR Implementation Surgical Site Infection Rates
Giannoula S. Tansarli,*†Lindsay LeBlanc,* Dianne B. Auld,‡and Kimberle C. Chapin†§
From the Departments of Pathology* and Infection Control,‡ Rhode Island Hospital, Providence; and the Departments of Pathology and Laboratory Medicine† and Medicine,§ Warren Alpert Medical School of Brown University, Providence, Rhode Island

Abstract
Nasal colonization with Staphylococcus aureus is a well-referenced risk factor for postoperative surgical site infections (SSIs). Our health care system that performs >40,000 surgeries per year assessed both the diagnostic accuracy of the BD MAX StaphSR assay (MAX StaphSR), a PCR-based test that detects and differentiates S. aureus and methicillin-resistant S. aureus (MRSA), compared with our standard of care culture and the subsequent clinical impact on SSIs 1 year after implementation. In addition, residual specimens were tested by broth-enriched culture. Performance parameters for all methods were determined using latent class analysis. Direct culture was the least sensitive for S. aureus (85.1%) and MRSA (76.7%), whereas the MAX StaphSR assay and broth-enriched culture had similar sensitivities (96.7%) for MRSA. Prospective assessment using MAX StaphSR during a 1-year, postimplementation period revealed a lower rate of SSIs per 100 targeted surgeries (0.3) compared with MRSA-only screening (1.10) and no screening (2.28) (P < 0.05 for StaphSR versus MRSA-only screening and StaphSR versus no testing). MRSA and methicillin-sensitive S. aureus SSIs occurred equally (n = 14 each). The MAX StaphSR assay provided accurate detection of both S. aureus and MRSA nasal colonization in presurgical patients, allowing infection prevention measures, including presurgical prophylaxis, to be implemented in a timely and consistent manner to avoid SSIs. (J Mol Diagn 2020, 22: 1063–1069; https://doi.org/10.1016/j.jmoldx.2020.05.003)
Nasal carriage with Staphylococcus aureus, including methicillin-resistant S. aureus (MRSA), is a major risk factor for surgical site infections (SSIs).1, 2, 3, 4 In particular, colonization with MRSA poses a high risk for MRSA-related SSIs and related complications.5,6 Approaches for preventing SSIs include presurgical screening on patients for colonization with S. aureus and MRSA followed by decolonization and/or antibiotic prophylaxis, as part of a comprehensive program.2, 7, 8, 9,10 The BD MAX StaphSR assay (MAX StaphSR; Becton, Dickinson and Company, BD Life Sciences—Integrated Diagnostic Systems, Sparks, MD) is a PCR-based assay that detects and differentiates S. aureus DNA and MRSA DNA from nasal swabs. Amplification targets include the thermostable S. aureus nuc gene, the mec right-extremity junction (MREJ), and the mecA/mecC methicillin resistance genes. Positive results for nuc and mecA/mecC are required for a test result of MRSA positive, whereas nuc or MREJ, in the absence of mecA/ mecC, are necessary for a test result of S. aureus positive, interpreted as methicillin-sensitive S. aureus (MSSA).
The objective of this study was to compare the MAX StaphSR assay and a standard direct culture method for screening of nasal colonization with S. aureus and MRSA in patients before surgery. A second objective was to investigate whether MAX StaphSR differentially affects the occurrence of SSIs caused by S. aureus in a multihospital setting compared with clinician-ordered screening for MRSA only (MRSA-SC; CHROMagar plating; Becton, Dickinson and Company, BD Life Sciences) or no screening.
Materials and Methods
Diagnostic Performance for Detection of S. aureus and MRSA
Presurgical nasal culture swab-based screening was performed, per standard protocol, with a Copan Dacron double swab (Copan Diagnostics, Murrieta, CA) between May 2015 and October 2015. Per the instructions of the package insert of the assay, both swabs were inserted simultaneously into each nostril. Multiple diagnostic approaches for S. aureus and MRSA were employed during study conduct.
Culture-Based Detection
For direct culture (standard of care), one swab was inoculated onto BBL Columbia Colistin Nalidixic Acid (CNA) with 5% Sheep Blood (Becton, Dickinson and Company, BD Life Sciences—Integrated Diagnostic Systems) for routine direct culture, and the plates were examined after 24 to 48 hours of incubation at 35oC. Presumptive S. aureus colonies were confirmed by catalase and coagulase testing, and methicillin susceptibility or resistance was determined using BBL Oxacillin Screen Agar (Becton, Dickinson and Company, BD Life Sciences) incubated at 24 hours at 35oC.11Direct culture report indicated the presence and/or absence of MRSA and/or S. aureus. For broth-enriched culture, 200 mL of the residual specimen buffer tube contents was transferred to a BBL Trypticase Soy Broth (Becton, Dickinson and Company, BD Life Sciences) with 6.5% sodium chloride for 24 to 48 hours of incubation at 35oC. If growth was observed, the BBL Trypticase Soy Broth with 6.5% sodium chloride was subcultured to CNA and processed, as described above, for direct culture.
MAX StaphSR
The second swab was tested by the MAX StaphSR assay on a BD MAX System (Becton, Dickinson and Company, BD Life Sciences—Integrated Diagnostic Systems) following the manufacturer’s instructions.
According to the package insert, for a specimen to be identified as MRSA positive with the BD MAX StaphSR assay, a positive result must be obtained for both MREJ and mecA or mecC; the nuc gene is not mandatory. The test uses three DNA targets: i) staphylococcal cassette chromosome mec–orfX right-extremity junction (MREJ), ii) thermostable nuclease of S. aureus (nuc), and iii) methicillin resistance (mecA/mecC ). In brief, i) detection of MREJ and mecA/ mecC is required for the result MRSA, ii) detection of nuc or MREJ without mecA/mecC is interpreted as positive for S. aureus, and iii) detection of nuc and mecA/mecC is interpreted as mixed MSSA and methicillin-resistant non–S. aureus, and detection of MREJ without mecA/ mecC is interpreted as S. aureus empty-cassette variant.
Discordant Analysis
Discordant results (algorithm for discordant analysis shown) (Table 1) between any of the three methods for a given specimen were tested further with various phenotypic methods, including catalase, Staphaurex (Thermo Fisher Scientific, Waltham, MA), BD BBL CHROMagar MRSA II (CHROMagar Chromogenic Media; Becton, Dickinson and Company, BD Life Sciences—Integrated Diagnostic Systems), and genotypic methods (mecA/C, nuc, and staphylococcal cassette chromosome mec/orfX PCR) using residual swab specimens, bacterial isolates, and residual specimen buffer tube contents, as needed. These tests were performed at Becton Dickinson Life Sciences (Québec, Canada), with technologists blinded to original results. To remain blinded, the technologists performed all of the tests described previously for each discordant result.
Table 1: Integration of Direct Culture, Enriched Culture, MAX StaphSR, and Discordant Analysis for Determination of a Discordant Result for Latent Class Analysis
| Direct culture | Enriched culture | MAX StaphSR | Discordant analysis result * | Discordant result |
|---|---|---|---|---|
| POS | POS | POS | n/a | POS |
| POS | NEG | NEG | POS | POS |
| NEG | POS | NEG | NEG | NEG |
| NEG | NEG | NEG | n/a | NEG |
| NEG | NEG | POS | POS | POS |
| NEG | NEG | POS | NEG | NEG |
*Discordant results between combined culture and MAX StaphSR resulted in additional testing, which included phenotypic methods (catalase, Staphaurex, and CHROMagar) and genotypic methods (mecA/C, nuc, and staphylococcal cassette chromosome mec/orfX PCR) using a specimen obtained from a residual swab (see Materials and Methods)
MAX StaphSR, BD MAX StaphSR assay; n/a, not applicable; NEG, negative result; POS, positive result.
Latent Class Analysis
Results from culture methods and MAX StaphSR were utilized to conduct latent class analysis (LCA) and determine performance for detection of S. aureus and MRSA.12,13 LCA is frequently used to assess diagnostic test accuracy when a gold standard for disease detection is not available. LCA assumes the existence of a latent categorical variable (e.g., presumptive true disease status) such that the observed response variables (e.g., various ratings on the disease status) are statistically, conditionally independent, given that variable (i.e., knowledge of one test result gives no information about other test results conditional on disease status). For statistical software, R version 3.12 (Microsoft, Redmond, WA) and SAS version 9.213 (SAS Institute, Cary, NC) were used to gather all test data points, assuming a probabilistic model. A contingency table was created to generate the latent class from the multiple tests employed in this study. Each test was then compared with the latent class to determine sensitivity and specificity. Only specimens with results available for all three tests (i.e., MAX StaphSR, direct culture, and broth-enriched culture) were used for the LCA. This study was reviewed and approved by the Lifespan—Rhode Island Hospital Institutional Review Board.
Surgical Site Infections
Preoperative screening for S. aureus with MAX StaphSR was implemented as the recommended presurgical screening test, to be performed 10 days to 2 weeks before surgery, for all patients in the hospital beginning February 14, 2018. Staphylococcus aureus screening was recommended to indicate preoperative infection prevention measures, which included Hibiclens bathing (Mölnlycke Health Care, Norcross, GA) and mupirocin nasal ointment before surgery for all patients who were identified as carriers of MSSA or MRSA. In addition, the results of MAX StaphSR were to be used for guidance of prophylactic antibiotic choice in the operating room in accordance with antimicrobial stewardship guidelines.14 The recommended surgical screening group included orthopedic, cardiac, neurosurgery, or any implantable device surgeries. Although the MAX StaphSR screen was recommended for presurgical screening as the preferred test, it was noted when tallying postimplementation data that providers were often ordering MRSA-only culture screen (MRSA-SC) using CHROMagar plating (our standard screen for medical admissions) or no preoperative screening test. Thus, although the original goal was to assess the performance of MAX StaphSR, whether MAX StaphSR surgical prescreening would impact SSI occurrence compared with MRSA-SC or no screening was also investigated. At this point, it is important to clarify that during the evaluation of the BD MAX StaphSR in our laboratory, standard of care included only direct culture with CNA agar for detection of S. aureus and MRSA. After evaluating the study results and before BD MAX StaphSR could be implemented in clinical practice, the decision was made to add CHROMagar in addition to CNA in an effort to increase the sensitivity of our culture method. Now, surgeons have the option to order a PCR (BD MAX StaphSR), an MRSA screen only (MRSA-SC), or no screening.
Screening methods and SSI data were collected prospectively for 12 consecutive months. The hospital infection prevention team tracked patients for SSI development for 3 months following the procedure date. The χ2 test was used to determine significant differences in the rate of SSIs between MAX StaphSR, MRSA-SC, and no screening. Odds ratios and accompanying CIs were calculated according to Altman15 (1991). For test of significance, a P value was calculated, as described previously by Sheskin16 (2004); a standard normal deviate was calculated as ln(OR)/SEM [ln(OR)], and the P value was the area of the normal distribution that falls outside the values of the normal deviate. P < 0.05 was selected to indicate statistical significance.
Table 2: Distribution of Staphylococcus aureus and MRSA Detection by MAX StaphSR, DC, and EC
| DC | EC | MAX StaphSR | LC* | Frequency | DC | EC | MAX StaphSR | LC* | Frequency |
|---|---|---|---|---|---|---|---|---|---|
| S. aureus detection | MRSA detection | ||||||||
| NEG | NEG | NEG | NEG | 772 | NEG | NEG | NEG | NEG | 1058 |
| NEG | NEG | POS | NEG | 58 | NEG | NEG | POS | NEG | 16 |
| NEG | POS | NEG | NEG | 6 | NEG | POS | NEG | NEG | 6 |
| NEG | POS | POS | POS | 41 | NEG | POS | POS | POS | 7 |
| POS | NEG | NEG | NEG | 5 | POS | NEG | NEG | NEG | 7 |
| POS | NEG | POS | POS | 12 | POS | NEG | POS | POS | 1 |
| POS | POS | NEG | POS | 5 | POS | POS | NEG | POS | 1 |
| POS | POS | POS | POS | 218 | POS | POS | POS | POS | 21 |
| Total | 1117 | Total | 1117 | ||||||
*LC is defined using an algorithm involving multiple test results to maximize the probability of correctly classifying infection status. Only specimens with available results for all three tests (DC, EC, and MAX StaphSR) were included in LC analysis.
DC, direct culture; EC enriched culture; LC, latent class; MAX StaphSR, BD MAX StaphSR assay; MRSA, methicillin-resistant S. aureus; NEG, negative result; POS, positive result
Table 3: Performance Characteristics of the MAX Staph SR, DC, and EC Compared with Latent Class Analysis Reference
| Variable | Test | Sensitivity, % ( n/total) | 95% CI, % | Specificity, % ( n/total) | 95% CI, % |
|---|---|---|---|---|---|
| Staphylococcus aureus | MAX StaphSR | 98.2 (271/276) | 95.8–99.2 | 93.1 (783/841) | 91.2–94.6 |
| DC | 85.1 (235/276) | 80.5–88.9 | 99.4 (836/841) | 98.6–99.7 | |
| EC | 95.7 (264/276) | 92.6–97.5 | 99.3 (835/841) | 98.5–99.7 | |
| MRSA | MAX StaphSR | 96.7 (29/30) | 83.3–99.4 | 98.5 (1071/1087) | 97.6–99.1 |
| DC | 76.7 (23/30) | 59.1–88.2 | 99.4 (1080/1087) | 98.7–99.7 | |
| EC | 96.7 (29/30) | 83.3–99.4 | 99.4 (1081/1087) | 98.8–99.7 |
DC, direct culture; EC, enriched culture; MAX StaphSR, BD MAX StaphSR assay; MRSA, methicillin-resistant S. aureus.
Results
MAX StaphSR versus Culture Method for Detection of S. aureus and MRSA
A total of 1124 paired nasal swab specimens were screened for S. aureus (i.e., both MSSA and MRSA) by direct culture, broth-enriched culture, and MAX StaphSR; seven specimens were excluded from analysis because of incomplete data. Of 1117 evaluable specimens, 218 (19.5%) were positive for S. aureus by all three methods, and 21 (1.9%) were MRSA positive (Table 2). For any S. aureus, 772 specimens (69.1%) had concordant results by all three methods, whereas 1058 specimens (94.7%) had concordant negative results by all three methods for MRSA. Direct culture yielded 210 MSSA positive results (18.8%) and 30 MRSA positive results (2.7%). Broth-enriched culture yielded 235 MSSA positive results (21.0%) and 35 MRSA positive results (3.1%). MAX StaphSR detected 284 specimens (25.4%) as S. aureus, with 45 (4.0%) identified as MRSA.
A total of 127 and 38 specimens yielded discordant S. aureus and MRSA results, respectively, across the three test methods (Table 2). Importantly, of the 14 specimens that were positive for MRSA by either culture method, but negative for MRSA by MAX StaphSR, 7 could not be confirmed as MRSA positive through discordant analysis by mecA/C and staphylococcal cassette chromosome mec/orfX alternate PCR testing. These seven specimens were considered false positive for MRSA by culture method. Therefore, LCA modeling was employed as the gold standard12 to preclude direct or broth-enriched culture as reference methods. Direct culture was the least sensitive method for S. aureus (85.1%) and MRSA (76.7%), whereas MAX StaphSR and broth-enriched culture had similar sensitivities (96.7%) for MRSA when analyzed by the LCA model (Table 3). MAX StaphSR demonstrated specificity similar to direct and broth-enriched culture methods for MRSA detection, with a slightly lower (approximately 6%) specificity for any S. aureus detection compared with the two culture methods. From the 16 specimens that were MRSA positive by only MAX StaphSR, 2 were confirmed as MRSA by PCR, 7 showed no growth, and 7 specimens had MSSA as determined by culture. Alternate PCR testing, as part of discordant analysis, determined that these latter seven MSSA isolates were mecA dropout, S.aureus-positive specimens, copositive for coagulase-negative Staphylococcus species harboring the mecA gene. Of the 58 specimens that were positive for S. aureus by MAX StaphSR only, 13 were confirmed as having S. aureus by nuc PCR testing. Analysis for the remaining specimens revealed a low bacterial load for S. aureus around the assay limit of detection (approximately 200 colony-forming units/swab). Per the package insert, the limit of detection of the assay, defined as the lowest concentration at which at least 95% of all replicates tested positive, ranged from 71 to 454 colony-forming units/swab for the detection of MRSA strains and from 174 to 211 colony-forming units/swab for the detection of MSSA strains.
Table 4: Data Involving Surgeries, SSIs, and Presurgical Screening between February 2018 and February 2019 at Rhode Island Hospital
| Screening | Data |
|---|---|
| Surgeries, N | 3388 |
| Prior screening, n (%) | |
| MAX StaphSR | 2050 (59.7) |
| MRSA-SC | 724 (21.1) |
| No testing | 614 (17.9) |
| SSIs, N | 28 |
| Pathogen, n | |
| Staphylococcus aureus | 14 |
| MRSA | 14 |
| Prior screening, n | |
| MAX StaphSR | 6 (2 positive*; 4 negative) |
| MRSA-SC | 8 (1 positive; 7 negative) |
| No testing | 14 |
| Rate of SSI, † % ( n/total) | |
| MAX StaphSR | 0.3 (6 ‡/2050) |
| MRSA-SC | 1.10 (8 §/724) |
| None | 2.28 (14 ¶/614) |
MAX StaphSR, BD MAX StaphSR assay; MRSA, methicillin-resistant S. aureus; MRSA-SC, methicillin-resistant S. aureus screening with CHROMagar; SSI, single-site infection.
*Two positive results during prior screening by MAX StaphSR consisted of one methicillin-sensitive S. aureus positive and one MRSA positive.
†SSI rate per 100 cases of respective surgery type.
‡Six SSIs consisted of five methicillin-sensitive S. aureus infections and one MRSA infection.
§Eight SSIs consisted of five methicillin-sensitive S. aureus infections and three MRSA infections.
¶Fourteen SSIs consisted of 10 methicillin-sensitive S. aureus infections and 4 MRSA infections.
Surgical Site Infections
Between February 2018 and February 2019, 3388 surgeries were performed within the study population. The presurgical screening process included a choice for clinicians between MAX StaphSR testing (n = 2050) or MRSA-SC (CHROMagar plating for MRSA growth; n = 724). Patients came to either a preoperative clinic site or a phlebotomy draw site to have provider orders performed. Specimens for MAX StaphSR testing were run daily, Monday to Friday, and results were provided consistently by 10:00 a.m. each morning to ensure that the presurgical team could follow up with patients on presurgical instructions. For MRSA-SC, specimens were prepared in batches and included with all MRSA culture screens performed for medical admissions. MRSA culture results were inconsistently reported to providers because of receipt time in the microbiology laboratory and batch read times by two different shifts. MRSA-only testing allowed for the appropriate preoperative preventive measures and vancomycin at the time of surgery; however, MRSA-only testing facilitated no preventive measures for MSSA-positive patients. An absence of presurgical screening allowed for no specification of infection risk and no specific implementation of prophylactic measures for patients. There were 614 cases considered high risk and appropriate for preoperative screening, which received no test screening.
During the 1-year assessment period, 28 cases of SSIs occurred, 14 of which were caused by MSSA and another 14 of which were caused by MRSA (Table 4). The 28 cases involved spinal fusion (n = 10), craniotomy (n = 8), laminectomy (n = 5), colon surgery (n = 2), knee prosthesis (n = 1), coronary artery bypass grafting (n = 1), and hip arthroplasty (n = 1) surgeries. Among the 28 patients who developed an SSI, 8 were screened by MRSA-SC and 6 by MAX StaphSR; no screening accounted for 14 patients. Only 3 of the 28 patients with an SSI were positive in the presurgical screening period (1 MRSA by MAX StaphSR, 1 for S. aureus by MAX StaphSR, and 1 MRSA by MRSA-SC). Eleven patients were negative on screening (four with MAX StaphSR and seven with MRSA-SC) but later developed an SSI (nine due to MSSA and two due to MRSA). All four of the negative screen results with MAX StaphSR were MSSA infections, and five of the seven patients who were negative at screening by MRSA-SC later developed an MSSA SSI. In 10 of the 14 patients with unknown status of colonization before surgery, the infection was caused by MRSA. Overall, MRSA SSIs were more common among patients who did not have a presurgical screening than those who had a screening test with either MAX StaphSR or MRSA-SC (71.4% versus 28.6%; P < 0.05). In addition, the SSI rate per 100 targeted surgeries, according to implemented screening method, was 0.24 for those screened with MAX StaphSR, 1.10 for MRSA-SC, and 2.28 for those who did not have any screening test (Table 4). The difference in SSI rate per 100 targeted surgeries between both MAX StaphSR and no testing, and MAX StaphSR and MRSA-SC, was statistically significant (P < 0.001 and P = 0.008, respectively). However, the SSI rates did not differ significantly between MRSA-SC and no testing.
Odds ratios were calculated to determine the association between screening method and SSI as an outcome in the no screening, MRSA-SC, and MAX StaphSR groups. No screening showed a significantly higher association with postoperative SSI compared with MAX StaphSR for any S. aureus infection and for MRSA infection; no screening did not have a significantly higher association with MSSA compared with MAX StaphSR (Figure 1). In addition, no screening had a higher association with MRSA compared with MRSA-SC screening. Although a trend was observed for a higher association with postoperative MRSA infection with MRSA-SC-based screening compared with MAX StaphSR, this was not statistically significant.
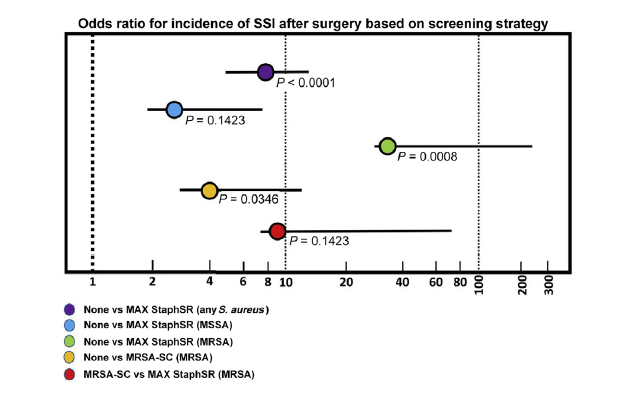
Figure 1: Odds ratios reflecting the association of screening approach with the incidence of postoperative Staphylococcus aureus—methicillin-sensitive S. aureus (MSSA)—and methicillin-resistant S. aureus (MRSA)-related single-site infections (SSIs). The three presurgical screening approaches [no screening, MRSA screening (CHROMagar; MRSA-SC), and BD MAX StaphSR assay (MAX StaphSR)] were conducted across a 1-year period of time. Postoperative surveillance was conducted for 3 months for any S. aureus-related SSI. For no screening compared with MAX StaphSR for any S. aureus, MSSA, and MRSA, the odds ratios were 7.9 (95% CI, 3.0–20.8; P < 0.0001), 2.7 (95% CI, 0.7–10.0; P = 0.1423), and 33.9 (95% CI, 4.3–265.35; P = 0.0008), respectively. For no screening compared with MRSA-SC for a MRSA-related SSI, the odds ratio was 4.0 (95% CI, 1.1–14.5; P = 0.0366). For MRSA-SC compared with MAX StaphSR for a MRSA-related SSI, the odds ratio was 8.6 (95% CI, 0.9–82.1; P = 0.0636).
Discussion
In this study, direct culture was the least sensitive method for detection of S. aureus and MRSA, whereas sensitivities for MRSA were similar with the MAX StaphSR PCR assay and broth-enriched culture. MAX StaphSR correctly identified more specimens with S. aureus than either culture method. The specificities for all three methods ranged from 93.1% to 99.4%. The lower specificity of the MAX StaphSR assay for S. aureus (93.1%) in this study is consistent with the assay performance in the product package insert. Possible explanations for false-positive MRSA results include cocolonization of the nares with an MSSA and coagulase-negative Staphylococcus species strain carrying the mecA gene, PCR detecting nucleic acid that does not differentiate viable from dead bacteria, and patients being on antibiotic therapy at the time of specimen collection, which could have affected recovery of bacterial isolates in culture.
The findings herein showing that the MAX StaphSR assay identified more S. aureus and MRSA than direct culture with CNA are similar to other studies that report an underestimation of S. aureus (MRSA and MSSA) carriage using standard direct culture methods.17,18 Although previous studies validated the assay for specimens collected with ESwab (Copan Diagnostics), showing excellent overall agreement with the culture,19,20 it is important to highlight that the current study was performed with Copan Dacron double swab, for which the assay is Food and Drug Administration approved. A key strength of this study was that all evaluated specimens underwent testing by three methods (direct culture, broth-enriched culture, and MAX StaphSR) and those with discordant results were tested by an array of phenotypic and genotypic methods. One advantage of using LCA modeling to determine test performance in this study was that it combined the results of direct culture, broth-enriched culture, and the MAX StaphSR assay to improve the classification of the disease status in an objective manner, compared with a single imperfect reference or composite of several imperfect references.12,13
The performance of the MAX StaphSR assay was presented to the Surgery Committee and subsequently implemented as the recommended preoperative screening test. A year after the assay was in place, the S. aureus-related SSI rate was determined to be approximately 5- and 10-fold higher for MRSA-SC (CHROMagar screen only) and no screening, respectively, in patients compared with those screened by MAX StaphSR. Odds ratio results showed a significantly greater association of no screening with MRSA-related SSIs compared with MRSA-SC and MAX StaphSR. Although MRSA-SC showed a significantly higher rate of overall SSIs compared with MAX StaphSR, the odds ratio of MRSA-SC to MAX StaphSR for MRSA- related SSIs (8.6) was not statistically significant (P = 0.0636). However, this analysis may not have been sufficiently powered to detect real differences between MRSA-SC and MAX StaphSR. Regardless, the fact that half of the S. aureus-related SSIs in this hospital population were caused by MSSA stresses the importance of screening preoperatively for both MRSA and MSSA. This is important for both risk awareness (risk of an SSI from any S. aureus is threefold higher for carriers in a hospital setting)21 and for appropriate antibiotic prophylaxis on the basis of screening results.
In terms of health care system performance measures, the MAX StaphSR assay allows better compliance with newly implemented hospital-wide preoperative guidelines established with surgery and our infectious diseases stewardship pharmacists. Specifically, the ability for targeted and appropriate use of the broad-spectrum antibiotic vancomycin was improved. For the preoperative center, the benefit over the nasal culture screen was the consistent reporting time of the MAX StaphSR results to the preoperative nursing service, which allowed a streamlined workflow and reporting to the electronic medical record for the anesthesiologists and surgeons.
Certain limitations are associated with this work. Limitations herein include the fact that this study involved LCA prevalence and test accuracy parameters that might have been better defined. Furthermore, the results for test accuracy obtained from LCA were generated on the basis of estimates and therefore are subject to a certain level of uncertainty although this is a common limitation for many statistical techniques.12 Another limitation involves the real- world data obtained concerning SSIs. The analyses used for SSI incidence over 1 year were performed using data associated with numerous independent variables (e.g., different surgery types, different times of year, and different composition of surgery teams) and therefore it is difficult to establish that the significant differences between MAX StaphSR and no screening are due solely to the coverage provided by PCR-based screening before surgery. Future studies are required to test this hypothesis and account for these types of confounding factors to clearly delineate the efficacy of PCR-based screening for S. aureus to prevent SSIs. Finally, these data were collected within a single hospital system, despite the inclusion of three hospital sites, which utilizes routine presurgical screening and prophylaxis as one measure to reduce SSIs, which may not be applicable to other sites.
In summary, the current data suggest that the BD MAX StaphSR assay provides accurate and rapid detection of S. aureus and MRSA colonization. Thus, MAX StaphSR could be successfully utilized as a screening tool to facilitate patient management, including both infection prevention measures and appropriate preoperative antibiotic prophylaxis, as part of a comprehensive SSI prevention strategy.
Author Contributions
All authors contributed to the interpretation of the data, critically revised the manuscript for important intellectual content, approved the final version to be published, and agree to be accountable for all aspects of the work.
References
- Abbas M, Aghayev E, Troillet N, Eisenring MC, Kuster SP, Widmer AF, Harbarth S, SwissNoso: Temporal trends and epidemiology of Staphylococcus aureus surgical site infection in the Swiss Surveillance Network: a cohort study. J Hosp Infect 2018, 98:118e126
https://www.sciencedirect.com/science/article/abs/pii/S0195670117305340 - Ban KA, Minei JP, Laronga C, Harbrecht BG, Jensen EH, Fry DE, Itani KMF, Dellinger EP, Ko CY, Duane TM: Executive summary of the American College of Surgeons/Surgical Infection Society surgical site infection guidelines—2016 update. Surg Infect (Larchmt) 2017, 18: 379e382
https://www.liebertpub.com/doi/10.1089/sur.2016.214 - Kalmeijer MD, van Nieuwland-Bollen E, Bogaers-Hofman D, de Baere GA: Nasal carriage of Staphylococcus aureus is a major risk factor for surgical-site infections in orthopedic surgery. Infect Control Hosp Epidemiol 2000, 21:319e323
https://www.cambridge.org/core/journals/infection-control-and-hospital-epidemiology/article/nasal-carriage-of-staphylococcus-aureus-is-a-major-risk-factor-for-surgicalsite-infections-in-orthopedic-surgery/F7844C48257F2D0646F65AFD88DD5092# - Levy PY, Ollivier M, Drancourt M, Raoult D, Argenson JN: Relation between nasal carriage of Staphylococcus aureus and surgical site infection in orthopedic surgery: the role of nasal contamination: a systematic literature review and meta-analysis. Orthop Traumatol Surg Res 2013, 99:645e651
https://www.sciencedirect.com/science/article/pii/S1877056813001461 - Allareddy V, Das A, Lee MK, Nalliah RP, Rampa S, Allareddy V, Rotta AT: Prevalence, predictors, and outcomes of methicillin-resistant Staphylococcus aureus infections in patients undergoing major surgical procedures in the United States: a population-based study. Am J Surg 2015, 210:59e67
https://www.sciencedirect.com/science/article/abs/pii/S0002961015000173 - Kalra L, Camacho F, Whitener CJ, Du P, Miller M, Zalonis C, Julian KG: Risk of methicillin-resistant Staphylococcus aureus surgical site infection in patients with nasal MRSA colonization. Am J Infect Control 2013, 41:1253e1257
https://www.sciencedirect.com/science/article/abs/pii/S0196655313009462 - Bode LG, Kluytmans JA, Wertheim HF, Bogaers D, Vandenbroucke- Grauls CM, Roosendaal R, Troelstra A, Box AT, Voss A, van der Tweel I, van Belkum A, Verbrugh HA, Vos MC: Preventing surgical-site infections in nasal carriers of Staphylococcus aureus. N Engl J Med 2010, 362:9e17
https://www.scopus.com/home.uri - Bode LG, van Rijen MM, Wertheim HF, Vandenbroucke-Grauls CM, Troelstra A, Voss A, Verbrugh HA, Vos MC, Kluytmans JA: Long-term mortality after rapid screening and decolonization of Staphylococcus aureus carriers: observational follow-up study of a randomized, placebo-controlled trial. Ann Surg 2016, 263:511e515
https://www.scopus.com/home.uri - Pofahl WE, Goettler CE, Ramsey KM, Cochran MK, Nobles DL, Rotondo MF: Active surveillance screening of MRSA and eradication of the carrier state decreases surgical-site infections caused by MRSA. J Am Coll Surg 2009, 208:981–986. discussion 986-988
https://www.sciencedirect.com/science/article/abs/pii/S1072751509001197 - Schweizer ML, Chiang HY, Septimus E, Moody J, Braun B, Hafner J, Ward MA, Hickok J, Perencevich EN, Diekema DJ, Richards CL, Cavanaugh JE, Perlin JB, Herwaldt LA: Association of a bundled intervention with surgical site infections among patients undergoing cardiac, hip, or knee surgery. JAMA 2015, 313:2162–2171
https://jamanetwork.com/journals/jama/fullarticle/2300601 - Tjalma WA, Depuydt CE: Cervical cancer screening: which HPV test should be used–L1 or E6/E7? Eur J Obstet Gynecol Reprod Biol 2013, 170:45–46
https://www.sciencedirect.com/science/article/abs/pii/S030121151300287X - Pepe MS, Janes H: Insights into latent class analysis of diagnostic test performance. Biostatistics 2007, 8:474–484
https://academic.oup.com/biostatistics/article/8/2/474/232752 - van Smeden M, Naaktgeboren CA, Reitsma JB, Moons KG, de Groot JA: Latent class models in diagnostic studies when there is no reference standard—a systematic review. Am J Epidemiol 2014, 179:423–431
https://academic.oup.com/aje/article/179/4/423/128491 - Bratzler DW, Dellinger EP, Olsen KM, Perl TM, Auwaerter PG, Bolon MK, Fish DN, Napolitano LM, Sawyer RG, Slain D, Steinberg JP, Weinstein RA: Clinical practice guidelines for antimicrobial prophylaxis in surgery. Am J Health Syst Pharm 2013, 70:195–283
https://academic.oup.com/ajhp/article-abstract/70/3/195/5112717?redirectedFrom=fulltext - Altman DG: Practical Statistics for Medical Research. London, UK: Chapman & Hall, 1991
https://scholar.google.com/scholar_lookup?title=Practical%20Statistics%20for%20Medical%20Research&author=D.G.%20Altman&publication_year=1991 - Sheskin DJ: Handbook of Parametric and Nonparametric Statistical Procedures. ed 3. Boca Raton, FL: Chapin & Hall/CRC, 2004
https://scholar.google.com/scholar_lookup?title=Handbook%20of%20Parametric%20and%20Nonparametric%20Statistical%20Procedures&author=D.J.%20Sheskin&publication_year=2004 - Bebko SP, Byers P, Green DM, Awad SS: Identification of methicillin-susceptible or methicillin-resistant Staphylococcus aureus carrier status preoperatively using polymerase chain reaction in patients undergoing elective surgery with hardware implantation. Infect Control Hosp Epidemiol 2015, 36:738–741
https://www.cambridge.org/core/journals/infection-control-and-hospital-epidemiology/article/identification-of-methicillinsusceptible-or-methicillinresistant-staphylococcus-aureus-carrier-status-preoperatively-using-polymerase-chain-reaction-in-patients-undergoing-elective-surgery-with-hardware-implantation/DDC3067EB3681987988C4FA5FEEA8B69 - Tsang STJ, McHugh MP, Guerendiain D, Gwynne PJ, Boyd J, Simpson A, Walsh TS, Laurenson IF, Templeton KE: Underestimation of Staphylococcus aureus (MRSA and MSSA) carriage associated with standard culturing techniques: one third of carriers missed. Bone Joint Res 2018, 7:79–84
https://www.scopus.com/home.uri - Silbert S, Kubasek C, Galambo F, Vendrone E, Widen R: Evaluation of BD max StaphSR and BD max MRSAXT assays using ESwab-collected specimens. J Clin Microbiol 2015, 53:2525–2529
https://www.scopus.com/home.uri - Silbert S, Gostnell A, Kubasek C, Widen R: Evaluation of the BD max StaphSR assay for detecting methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-susceptible S. aureus (MSSA) in ESwab-collected wound samples. J Clin Microbiol 2017, 55:2865–2867
https://www.scopus.com/home.uri - Wertheim HF, Vos MC, Ott A, van Belkum A, Voss A, Kluytmans JA, van Keulen PH, Vandenbroucke-Grauls CM, Meester MH, Verbrugh HA: Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 2004, 364:703–705
https://www.sciencedirect.com/science/article/abs/pii/S0140673604168979
Supported in part by Becton, Dickinson and Company, BD Life Sciences—Integrated Diagnostic Systems (K.C.C.).
Disclosures: The funders had no role in the planning or execution of the experiments included in this manuscript. They also had no role in the presentation of results or writing of this manuscript.
Originally published in The Journal of Molecular Diagnostics, Vol. 22, No. 8, August 2020.
Copyright© 2020 Association for Molecular Pathology and American Society for Investigative Pathology. Published by Elsevier Inc.
This is an OPEN ACCESS article under the CC BY-NC-ND license http://creativecommons.org/licenses/by-nc-nd/4.0/