Detecting Phenol in Nucleic Acid Samples with the Nanodrop One Spectrophotometer
Sample evaluation is a key step in many DNA- and RNA-based workflows since the success of an experiment is partially dependent on sample quality. The Thermo Scientific™ NanoDrop™ One Microvolume UV-Vis Spectrophotometer enables research scientists to accurately qualify and quantify nucleic acid samples. Built with the novel Acclaro Sample Intelligence technology, the NanoDrop One instrument detects common contaminants in nucleic acid samples. This data is valuable for troubleshooting problematic extractions and enables scientists to make more informed decisions about the suitability of samples for their experiments.
Purity Ratios: Only Part of the Story
DNA and RNA quantification, typically done by measuring absorbance at 260nm, is a crucial step for many molecular techniques performed in life sciences research. Absorbance at this wavelength (also known as A260) is proportional to the amount of nucleic acid in the sample; the nucleic acid concentration can be calculated using the Beer-Lambert law.
In addition to the concentration, scientists also want to know the purity level of their samples. The A260 value can be affected by the presence of substances that absorb in the same UV range as nucleic acids. Residual chemicals from nucleic acid extraction processes (such as guanidine salts or phenol) or cell components that co-extract with the nucleic acids (such as protein) can artificially inflate the A260 value.
To eliminate the effects of these contaminants, scientists may use the absorbance spectra to calculate purity ratios: A260/A280 and A260/A230.
Figure 1 Acclaro Contaminant ID identifies possible contaminants in a sample.
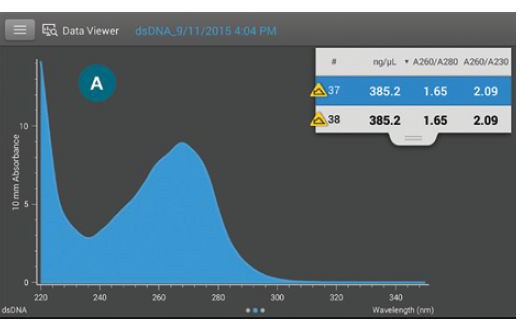
1A) Measurement screen: the yellow icon indicates that the Acclaro algorithms have detected a potential contaminant in this dsDNA sample.
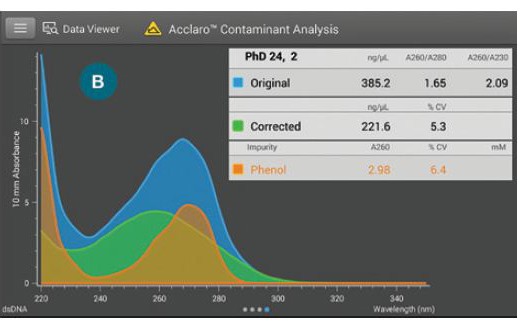
1B) Contaminant analysis screen: displays the comparison of original absorbance spectrum (DNA plus phenol, blue), the corrected spectrum (original minus contaminant, green), the contaminant spectrum (orange), and the original and corrected DNA concentrations.
NanoDrop instruments routinely include purity ratios with reported results. DNA samples with purity ratios that range from 1.8 to 2.0 are typically considered pure. Although purity ratios are very useful and informative, they do not always provide a complete picture of sample purity.
Contaminant Identification
A new Acclaro Contaminant Identification (ID) feature has been added to the NanoDrop One Spectrophotometer to detect protein, phenol and guanidine-salt contaminants in nucleic acid samples.
During sample measurement, the user is alerted in real time by a yellow contaminant ID icon (Figure 1A), which appears next to the sample number on the NanoDrop One touchscreen. Tapping the icon will reveal the full details of the Acclaro contaminant analysis (Figure 1B). The screen displays the deconvolved spectra, the contaminant contribution to the overall A260, and the corrected DNA concentration of the sample (Figure 1B). The corrected concentration is obtained by subtracting the contaminant A260 contribution from the original result.
| Mixture | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| Phenol content (ppm) | 0.00 | 18.75 | 37.50 | 75.00 | 150.00 | 300.00 | 600.00 | 1200.00 | 1600.00 |
| Original (Uncorrected) DNA conc. ng/µL | 245.75 | 253.50 | 264.34 | 284.75 | 318.35 | 387.55 | 523.59 | 851.29 | 1041.50 |
| Corrected DNA conc. ng/µL | *240.56 | *238.67 | 240.61 | 243.26 | 244.38 | 242.51 | 245.26 | 265.96 | 265.45 |
| Corrected DNA conc. ng/µL Std dev | 0.61 | 0.95 | 0.79 | 1.24 | 0.76 | 0.85 | 0.87 | 5.47 | 3.26 |
| 260/280 Purity Ratio | 1.89 | 1.85 | 1.83 | 1.79 | 1.75 | 1.70 | 1.71 | 1.53 | 1.54 |
| 260/230 Purity Ratio | 2.43 | 2.38 | 2.32 | 2.27 | 2.19 | 2.11 | 2.08 | 1.89 | 1.94 |
| Acclaro Flag | No Flag | No Flag | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ |
Table 1 The DNA concentration of each mixture (2-9) and the DNA-only control (1) were determined with the NanoDrop One spectrophotometer using the dsDNA application. The corrected DNA concentration for mixtures 3 through 9 was obtained directly from the Acclaro contaminant analysis screen. The Acclaro Contaminant ID icon (♦ ) denotes mixtures that have levels of phenol contamination high enough to trigger an Acclaro result.
Acclaro Detects Phenol, Corrects Concentrations
Phenol extraction is an established and trusted method for separating proteins from nucleic acids and is the basis for various nucleic acid extraction kits. Most extraction kits use varying formulations of phenol, guanidinium thiocyanate, and chloroform to lyse cells and denature proteins. Traces of these reagents (including phenol and guanidine salts) can be found in purified nucleic acid samples and may affect downstream experiments. The presence of phenol can prompt extensive troubleshooting since it can denature proteins and affect subsequent enzymatic steps. The NanoDrop Technical Support Group frequently receives calls that pertain to problems with extraction reagent contaminants.
Table 1 shows data collected on the NanoDrop One spectrophotometer using the dsDNA application.
Sample 1 is pure DNA in Tris-EDTA buffer. Samples 2-9 have been spiked with increasing concentrations of phenol.
This data illustrates that the Acclaro Contaminant ID feature:
- Accurately identifies phenol contamination present in nucleic acid samples
- Accurately calculates the amount of phenol contamination
- Provides corrected nucleic acid concentration values that are within 10% of the actual DNA concentration
These data also illustrate that increasing amounts of phenol contamination have minimal effect on the purity ratios.
A DNA sample with a phenol contamination of 600ppm has an A260/A230 ratio of 2.08, values that are within the generally accepted range for “pure” nucleic acid samples. However, the DNA concentration is generally off by greater than two-fold with this level of phenol contamination. These results demonstrate that one cannot rely solely on purity ratios to assess the purity of nucleic acid samples. Using the Acclaro algorithms, the NanoDrop One instrument can detect phenol contamination and provide corrected concentrations that overcome the purity ratio limitation. This critical information helps scientists decide if the sample can still be used or if it must be re-purified.

